Research
Under the menues below, you will find short deascriptions of our main research directions, together with pointers to key papers and collabiorators. For a complete list of papers please see our Publications page .
CRISPR/RNG core targeting
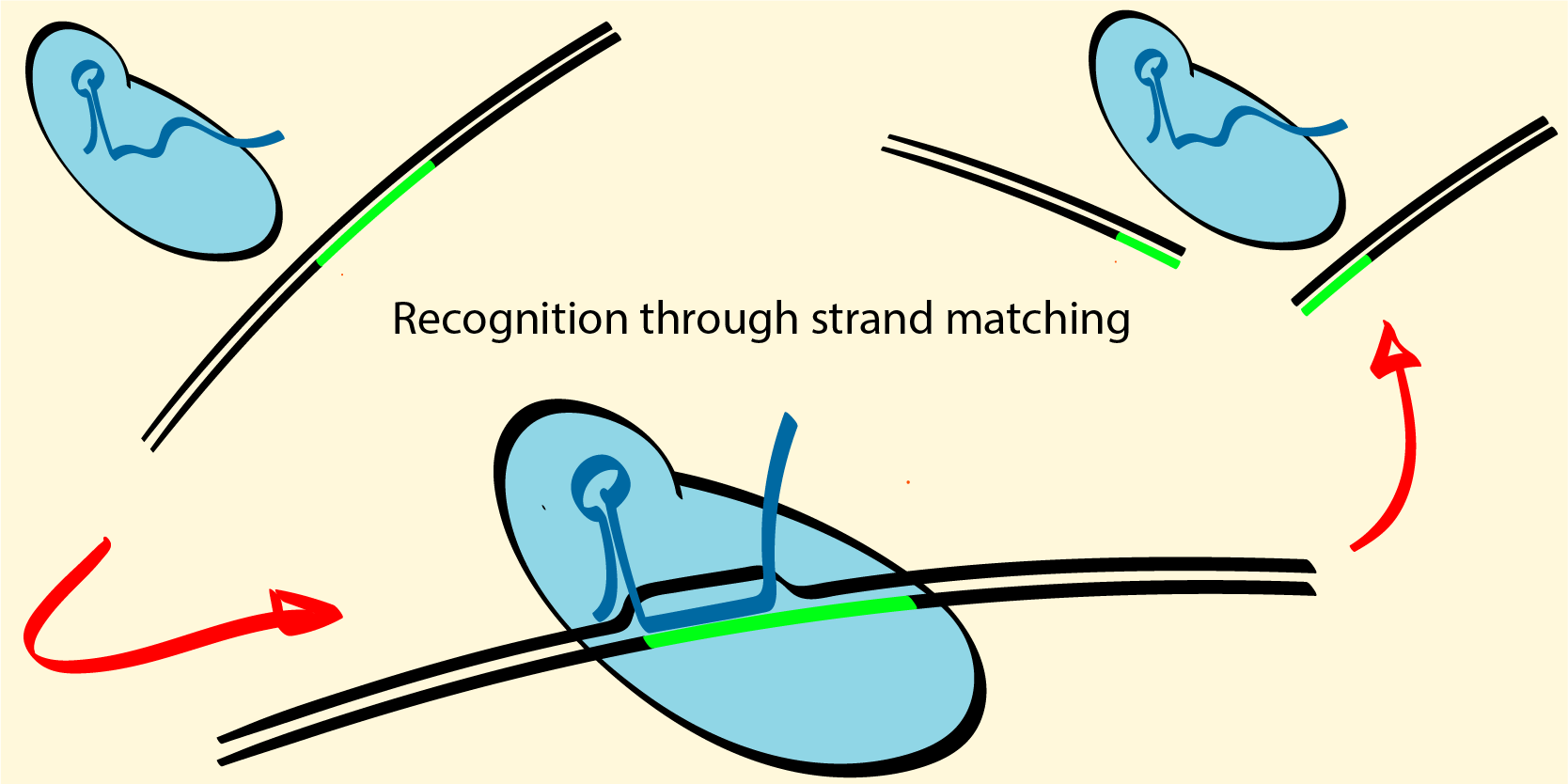
CRISPR based gene-expression and editing tools have revolutionized molecular biology and genetic engineering in animals and plants. Due to the extream precision requirments for human theraputics, CRISPR technologies are still not broadly used to treat genetic disorders. By modelling CRISPR based technologies in mechansitic terms, we characterize, predict, and help impove them.
We are working on a computational pipline capable of characterizing novel molecules and predict their core activity based on a very limited but targeted in vitro experimnents. Using a limited set of targeted experiments is possible due to the adaptability and limited complexity of physics-based models; it is desireable as it is inherently efficnet and adaptable to various situations, and because it leaves a lot of room for validation in a field where safety is paramount.
Selected publications
Collaborators
Other
Major contributors from our group
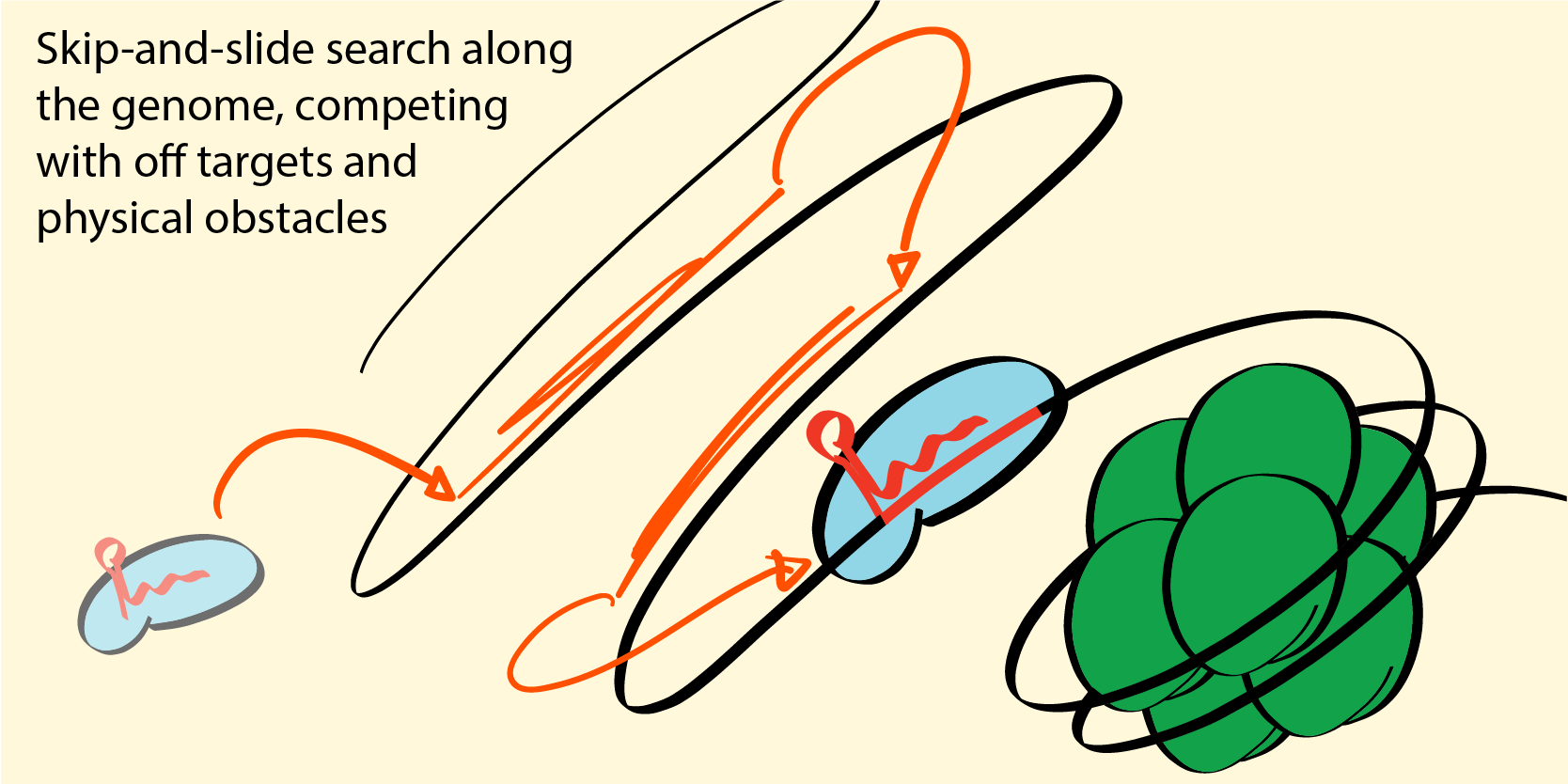
Before a target can be cleaved/bound, it needs to be found among a see of potential off-targets (6 billion in the human genome). For our training of the core targeting model on in vitro training data, protein concentrations are bufferded, and the presence or absence of competing sequences does not effect the outcome. In a cellular context, the situation is different: the acitve molecules are delivered or produced dynamically, copy number are relatively low, and the search happens in the precense of obstacles and billions of potential off-targets--many of which are close matches to the intended target.
As our core model of the targeting process is a physical model that yields quantitative predictions and not a simple ranking, the cellular context can be modelled and taken into accont. This context can be delivery method, copy numbers, chromatin state, and what individual genome is being targeted. Understanding the influence of the genome on the editing process will be crucial for bringing gene editing into personalized theraputics.
Selected publications
Collaborators
Major contributors in our group
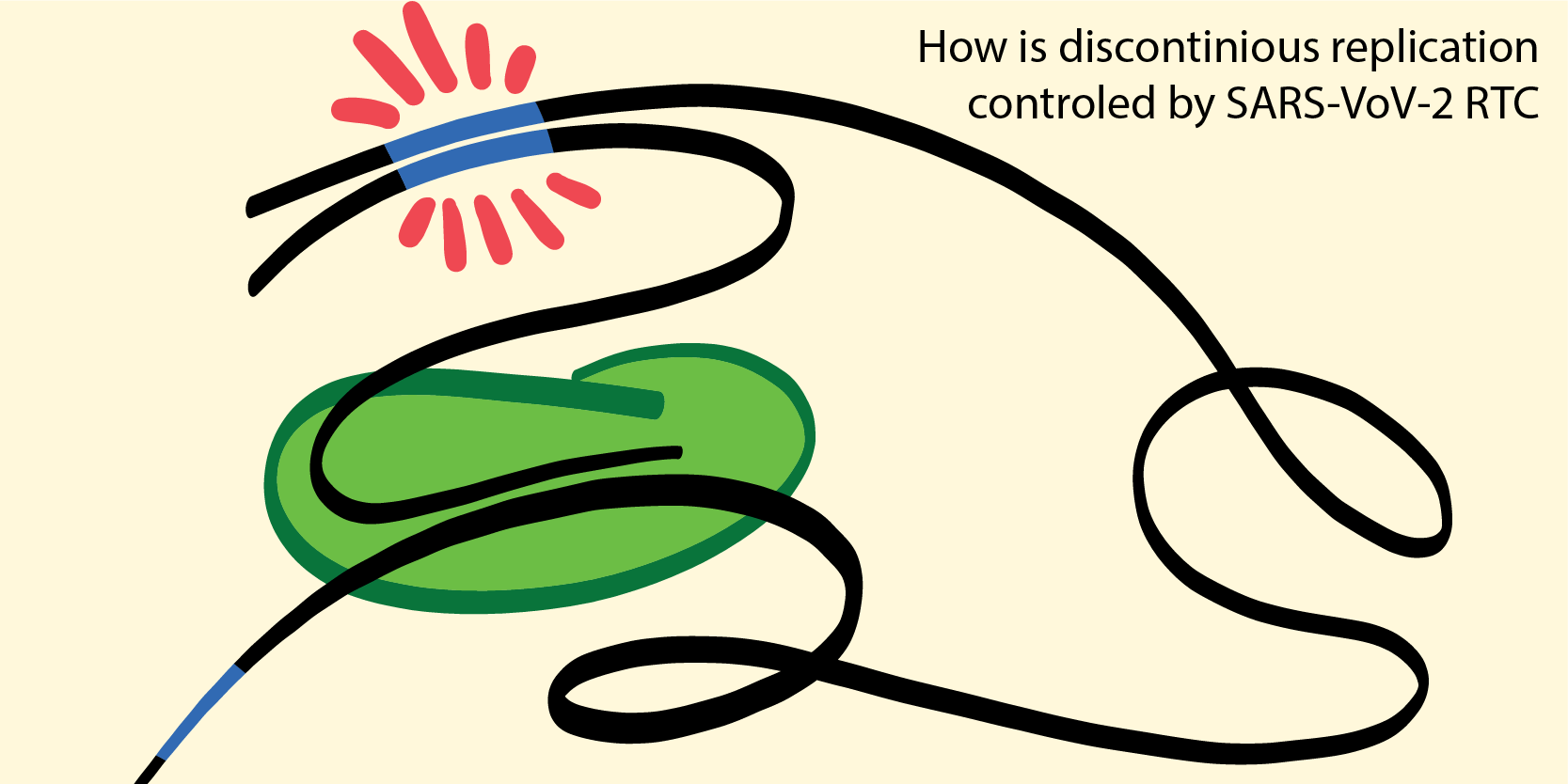
Viral pandemics are a threat to both the health and the economic wellbeing of societies. Many viruses also evolve quickly, bypassing vaccines over time. By targeting vital and highly conserved part of a virus, lasting vacciens and treatments might be found. Our efforts focus on mechansitically undersrtaning the viral replication process, and how it is impacted by context. Together with collaborators, we work on how various viral replication machines react to the introdcution of anti-viral drugs (nucleotide analogs). We presently exploring the role of specific seqeunces in regulating the viral replication process in SARS CoV-2.
Selected publications
Collaborators
Major contributors from our group
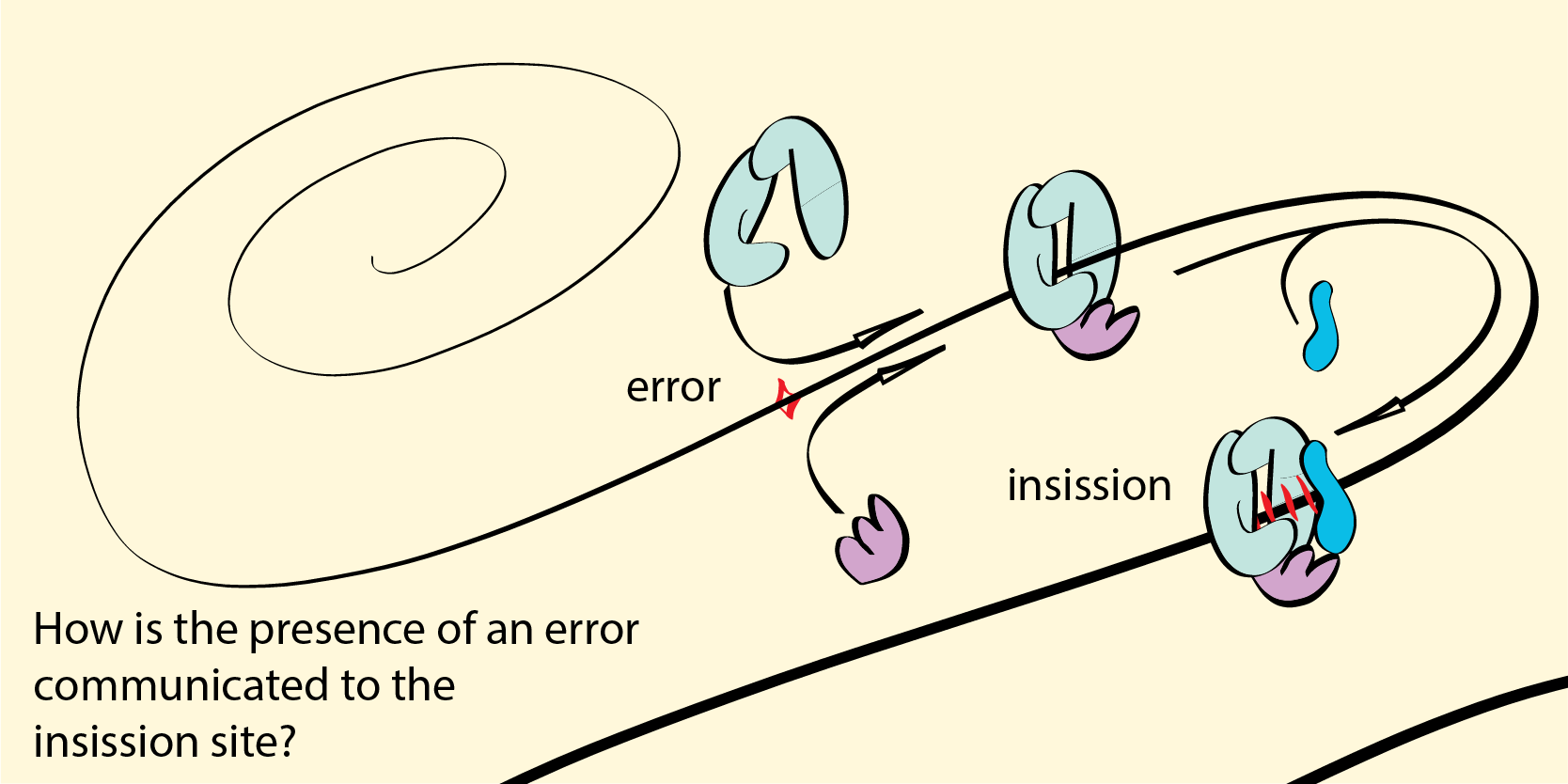
Errors are continiously made during the DNA replication process. To ensure genetic integrity, the cell has found ways of correcting such errors before the fitness of the individual is impacted and the reproductive success is decreased. The catching of errors is done either though proofreading in the replication process itself, or by detecting mismatches in a newly synthesized DNA strand and correcting them. In the latter case, ones an error is detected, a whole coreographed dance of molecules is needed to initiate the homology directed repair pathway. We study the initial steps of mismatch repair, trying to establsh the method by which the presence of an error is communicated from the site of the error to the site of the initial insission.